
Environmental monitoring has become an important “best practice” in cannabis production. Cannabis is a high value crop. Anything that contributes to preventing testing failure must be done to ensure crop protection, otherwise brands, companies, products and investors can be subjected to great losses.
Rapidly expanding cannabis regulations around the world are more frequently requiring higher and higher levels of sophisticated microbial contamination testing, which can be expensive. In reality, depending on regulations, cannabis products being tested for microbial contamination today are typically submitted without a high level of confidence that they will emerge compliant with existing regulations. This lack of confidence frequently leads to a harsh reality – risk of product failure.
When this high value product fails microbial testing, the product cannot be sold – leading to investment losses, grower losses, processor losses, manufacturing losses and less product on dispensary shelves that is safe. Most cannabis growers become keenly aware of the need for environmental testing after the first loss and in the search for prevention in the future.
We offer this paper with solutions and scientific data to solve this vexing problem for cannabis growers. The solution is to monitor the relevant areas within grows, processing/manufacturing sites and dispensaries for the presence of various microbes. In the past, this has been challenging in terms of time and expense to be able to use traditional culture methods for environmental monitoring. Utilizing molecular methods which can definitively detect and identify the species in a fraction of time and cost now makes monitoring a practical reality. The producers can diligently detect contamination issues before harvesting and processing product which permits producers to mitigate any contamination issues prior to submission of products to testing labs.
This article will describe the advantages of one particular molecular technology with the capability to perform this type of routine monitoring in a timely and cost effective manner and its advantages over gold standard plating methods as well as other molecular technologies.
Introduction
A recent report stated that in the first two months of the California regulations going into effect this last July, there were 114 batches of products rejected due to unacceptable microbial contamination (Blood, 2018). If the regulations change to include more microbial species as pushed for by at least one testing company, Cannalysis (Blood, 2018), then it is likely that the number of rejected lots of products will rise.
Most facilities which cultivate and/or process cannabis do so in a blind manner with respect to knowing if microbial contamination is present. These facilities sometimes implement aseptic processes to control microbial presence within the facility’s controlled environment. These processes include routine cleaning programs, and contamination control planning.
In the pharmaceutical industry, manufacturing facilities are required to routinely monitor various predetermined areas within it to collect samples and determine the kinds and levels of microbial contamination and establish alert and action levels (Sutton, 2010). Certainly, in the case of medicinal cannabis products, the standards for aseptic processing should mimic those for GMP-level manufacturing of pharmaceuticals.
Additionally, another approach to microbial risk mitigation that addresses preventative spread of microbes is outlined in the Hazard Analysis Critical Control Point (HACCP) and is sourced by the FDA as “a management system in which food and safety is addressed through the analysis and control of biological, chemical and physical hazards from raw material production, procurement and handling, to manufacturing, distribution and consumption of the finished product” (FDA 2018).
The main goal of HACCP is to avoid hazardous contamination (pathogenic microbes, chemical and others). This is accomplished through the implementation of preventative measures that aim to avoid contamination while maintaining procedural records for personnel to follow (SOP’s) thus resulting in optimal quality of product for the consumer and loss avoidance for the manufacturer.
Environmental monitoring of microbes involves the sampling from a number of different environments including surfaces, water and air. This article will focus on surface samples but air and water samples employ the same general approaches, though some of the technologies for collection may differ according to the sample type.
The general approach involves the following major steps (Jansen, 2014):
- Define a sampling plan including sampling locations and scheduling
- Collect samples
- Detect microbes in samples
- Report results that are reviewed and approved
- Implement mitigation actions, if necessary
- If mitigation actions are taken, collecting a new set of samples for follow-up analyses may be required to determine efficacy of those actions
For protecting consumers of cannabis-based products in states and countries where it is legal to possess and use cannabis, many companies have implemented regulations which limit the types and numbers of microbial species present in a specified amount of sample. These products are often submitted to license testing laboratories without knowledge of the likelihood of being compliant with the regulations. This is typically due to the lack of resources at the cultivation or processing site to perform microbial testing prior to the release of the product lot for official testing.
One could pretest their product prior to official submission of the lot which can determine whether a product is contaminated with unacceptable levels of one or more microbes specified by the regulations of that area. However, this approach is not conducive towards identifying the root cause or source of the contamination and thus could remain as a chronic issue for the site and subsequent products it is producing. Such situations can cause a company tremendous revenue loss and could jeopardize its existence, much less overall profitability.
The traditional “gold standard” for detecting microbial contamination has included the use of culture, microscopy, immunoassays and serology. Cannabis testing has mostly been paralleling the food safety testing standards which primarily utilizes culture-based methods for at least first order microbial contamination detection.
Environmental monitoring should utilize “SMART” tests to make the effective for cannabis growers and processors. The SMART tests are defined as:
- Simple – straightforward and easy to use with facile interpretation and reporting of test results
- Money-conscious – overall low cost of testing which encompasses not only reagents or kit, but also dedicated equipment, facilities and specially trained laboratory staff to run the tests
- Accountability - provide actionable results to understand location and potential sources of contamination which hold accountable the individuals and processes responsible for cleanliness of the affected area(s) within the facility
- Reliability – this is paramount to instill confidence that the results are accurate, informative and truly actionable
- Timely – early detection is necessary to implement mitigation actions early to prevent systemic contamination of the entire facility and product failures
Culture methods are suboptimal for rapid turnaround of results. Bacterial cultures take a minimum of 24 hours and can take several days longer if a slow growing species needs to be detected or if verification is required for organisms such as Salmonella. Fungal cultures inherently take more time due to their relatively slow growth rates compared to faster growing bacteria like E. coli. In addition, to identify species, it takes a trained microbiologist to identify species based on colony and cellular morphologies, as well as confirmatory testing.
Since the discovery of the structure of DNA three quarters of a century ago, genetic and genomic research has been explosive and have enabled greater and higher resolution understanding of species based on their genetic makeup. There are signature DNA sequences which permit highly accurate identification of species using a variety of techniques.
In essence, those specific DNA sequences represent truth in terms of whether an organism is present or not in a specimen. The methods available to identify species have become highly accurate and much more rapid, especially after the discovery of the polymerase chain reaction (PCR). PCR is an elegant and powerful technology which can amplify copies of specific segments of DNA in an exponential manner. Since its discovery, variations of PCR have greatly increased the types of applications possible for rapid genetic analysis of samples. One method which has taken PCR to a new level by enabling high density and parallel processing of DNA fragments for analysis is that of microarrays.
This paper will examine the potential for molecular methods, microarrays in particular, to be a SMART solution for environmental monitoring. We will examine the benefits of microarrays over culture for environmental monitoring.
Molecular assays comparative
With the emergence of molecular based assays, there have been several technological advancements that have introduced high levels of specificity and sensitivity in the area of pathogenic detection for food and agriculture safety. The primary method utilized in molecular based assays is polymerase chain reaction (PCR). PCR utilizes targeted primers coupled with a mastermix of distinct ions that amplifies the DNA analyte of interest with the use of a thermocycler.
Real time PCR (qPCR) allows amplification of the DNA analyte to be observed as it occurs. Amplification is detectable as an increasing fluorescent intensity signal crossing a threshold value or Cq (Kralik & Ricchi 2017). This process for microbial pathogen detection and identification is accomplished through the use of fluorescently labeled probes that are specific to the analyte of interest.
While qPCR holds a high level of sensitivity and specificity for its process in pathogenic detection, this technology cannot be used to multiplex or test for multiple pathogens simultaneously. Fluorescently labeled probe solutions must be purchased for each analyte being tested. This not only requires more laboratory preparation time but also introduces high costs to larger facilities that require more testing or high throughput.
Additionally, pathogenic detection is limited with this molecular method due to its lack of internal standard for absolute amplification comparison (Kralik & Ricchi 2017).
Next generation sequencing technology (NGS) is a significant advancement in molecular technology with the optimal level of specificity obtained by sequencing targeted microbial phylogenetic marker genes. This also offers the advantage of discovering many more novel microbes than with other molecular technologies (Frey & Bishop-Lilly 2015). Additionally, NGS holds inherent multiplexing. Due to its complexity however, NGS is the most costly microbe detection platform and requires a higher level of expertise and knowledge of molecular assays than any of the other platforms discussed.
An established molecular platform that is currently being utilized for pathogen detection is that of microarrays. Microarrays are small single stranded probes of DNA attached to a solid substrate (McLoughlin 2011). Similar to most molecular based methodology, microarrays utilize the amplified product of PCR thus making it a PCR based assay.
A major advantage with microarrays is that they can be vastly multiplexed with ease and can be designed to contain internal control standards that offer a higher standard of quality control especially in regulated environments. This offers cost savings as well as time savings (most microarrays offer less than one day turnaround time for results). Another advantage of microarrays are the ability to detect novel pathogens or those with heterogeneic variants due to its multiplexing capabilities (McLoughlin 2011). These advantages offer a significant cost reduction for customers that require a higher throughput of samples that are necessary to ensure pathogenic free facilities.
Experimental
For environmental monitoring of surfaces within a facility utilizing a molecular, microarray-based system, the order of major steps in the process starting with sample collection is below:
- Collect and secure sample
- Transport sample to laboratory
- Collect cell pellet
- Lyse cells
- Amplify specific DNA sequences using PCR
- Hybridize PCR products to probes on microarray
- Wash off non-specifically bound DNA
- Image microarrays with a fluorescence scanner
- Analyze data and issue report
Sample collection from innate surfaces can be accomplished in several different ways. The most common method is to take a swab (Thomas Scientific 1216P21; Mfr. No. 25-83004 in Butterfield’s Solution) that is pre-wetted and swabbing approximately one square foot by swabbing back and forth first horizontally and then vertically. The swab is then inserted into the original tube it was taken from and secured for transportation to the laboratory.
The tube with the swab is process in the lab in preparation for polymerase chain reaction (PCR) as follows using components from PathogenDx EnviroX Screening Kit (part no. ES-001):
- The swab in the tube with the transport medium is vortexed for 10 seconds.
- The fluid is transferred to a 1.5 ml microfuge tube centrifuged at low speed 800 x g for 3 minutes to remove large particulates. The supernatant is transferred to a new 1.5 ml microfuge tube.
- The tube is centrifuged at high speed (14,000 x g) for 3 minutes to pellet cells. As much supernatant is removed being careful not to disturb the cell pellet.
- Initial lysis buffer added then heat for 10 minutes at 95°C.
- To lysed cell material, add 50 µl Sample Buffer with sample digestion components already mixed in the buffer. Vortex for 10 seconds to resuspend the cells.
- Heat at 55°C for 45 minutes.
- Vortex for 10 seconds then heat 95°C for 15 minutes.
- Samples are now ready for PCR.
The DNA from the lysates (no DNA purification required) are then amplified using a proprietary two stage PCR protocol starting with loci enhancement and followed by labeling of the PCR amplification products. There are two sets of PCRs with one that is dedicated for bacterial species detection and the other is for fungal species detection. The loci enhancement PCR utilizes primers specific to either the 16S rDNA for the bacterial amplification or the ITS2 region for fungal amplification.
The labeling PCR uses a nested primer approach for both the bacterial and fungal amplifications in which one primer is labeled with a fluorescent dye (Cy3) which corresponds to the complementary strand of the probes on the microarray. The organisms represented on the microarrays are listed in Table 1.

After the PCRs are complete, these amplifications are then prepared for hybridization to the probes on the microarray. The basic protocol for hybridization and post-hybridization processes are described below:
- Place slides with microarrays into the hybridization chamber which has a moistened paper towel at the bottom of the chamber to create a high humidity environment. There are twelve separate microarrays, each in their own well, per slide. Each can be used for analyzing a single sample that is for either bacterial or fungal detection.
- Deposit 75 µl sterile molecular biology grade water to each well and wait for 5 minutes before aspirating the water from the slides.
- Add 50 µl Pre-hybridization Buffer to each well on the slide and leave on slide for 5 minutes at room temperature.
- Centrifuge the plate containing the post PCR product to bring all of the fluid to the bottom of the tube or well.
- Add 18 µl of Hybridization Buffer to each Labeling PCR and mix by pipetting up and down several times.
- Add 50 µl of the Hybridization Cocktail for each sample PCR to the designated well on the slide. Ensure that sample ID and location on the slide are recorded.
- Close Hybridization Chamber lid and allow samples to hybridize for 30 minutes at room temperature.
- Aspirate the Hybridization Cocktail from each well and then wash by adding 75 µl Wash Buffer and aspirating the solution off immediately after.
- Add 75 µl of Wash Buffer to each well and then leave at room temperature for 10 minutes and then aspirate.
- Final wash is adding 75 µl of Wash Buffer and immediately aspirate. This is repeated one more time.
- Slides are then dried by centrifuging in a Labnet Reusable Laboratory Micro Array Slide Spinner (part no. C-1303T) for 30 seconds.
- These slides are then placed into the slide holder for the PathogenDx Slide Scanner (part no. Slide-001) and then scanned.
- The scanned images are then analyzed using the Augury©™ analysis program.
Augury determines whether the samples are positive or negative for the presence of species represented in the microarray. Any positive call is based on whether the signal for that particular probe is above a threshold level. This is done on an individual probe basis since some probes have inherently higher general background signal than other probes.
A pilot study involved a facility in Arizona which was multifunctional having grow, processing and dispensary areas within the same building. This study examined the presence of bacterial and fungal species within the grow and processing areas only. In this study, instead of swabs, 2” x 2” adhesive lift tabs (Arrowhead Forensics, part no. A-2802LT) were used to collect samples from specific sites within the facility. These tabs were brought to PathogenDx for processing for microarray analysis.
Results
The collected adhesive lift tabs were processed with the PathogenDx environmental assay protocol as described. Numerical values for the probes on the arrays are shown in Figure 1 and a summary of the outcomes based on signals above the threshold levels are shown in Table 2. Weak positives below 10,000 relative fluorescence units (RFU) are highlighted in yellow and the strong positives above 10,000 RFU are shown with red highlighting.
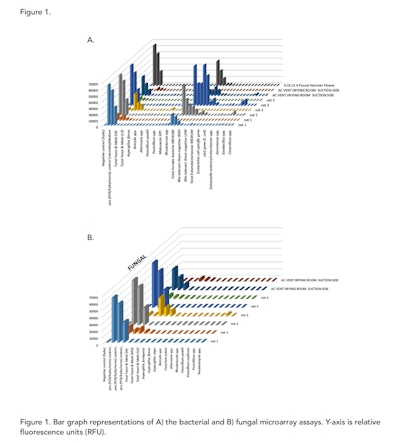
All samples except for the AC Duct Return in the Veg room had at least low levels yeast and mold and high levels of aerobic bacteria. Yet for the fungal assays, there were no individual species were identified which is not unexpected. On the bacterial side, there were several species which were identified at high levels, including Aeromonas spp. and Pseudomonas spp. These two species did cause concern for the operators of the facility that was tested and they did follow up with mitigation plans to “super” clean those areas. Follow up environmental monitoring was not performed in this case.
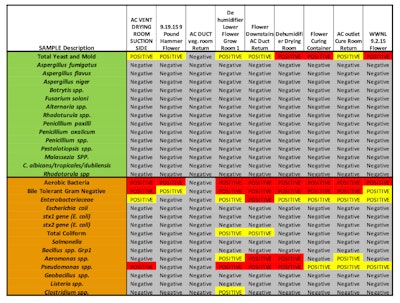 Table 2. Summary of results from microarray bacterial and fungal assays for Arizona pilot study
Table 2. Summary of results from microarray bacterial and fungal assays for Arizona pilot study
Additional study
A recent May 2019 study demonstrates the sensitivity and specificity of the EnviroX assay when compared to the standard plating method of pathogen detection for preventative environmental monitoring (PEM). This study was designed by a microbiologist and utilized optimal media for the growth of bacteria (tryptic soy agar (TSA)) and fungi (Sabouraud-dextrose agar (SDA) with chloramphenicol). The TSA plates were incubated at 25˚ C and the SDA was incubated at 25˚ C for 6 days. Concentrations plated were the neat, 1:10 and 1:100.
Areas that were presumed clean by the facility were swabbed and processed both with the plating method described above as well as with the EnviroX assay. Average times for both methods (from initial swab time) are summarized in Figure 2.
 Figure 2. Time comparison for plating vs EnviroX microarray pathogen detection system.
Figure 2. Time comparison for plating vs EnviroX microarray pathogen detection system.
Table 3 summarizes the results of plating vs EnviroX. While plating not only took more time, it also is subjective in speciation according to the expertise of the microbiologist. The EnviroX assay was able to not only classify bacterial contaminants based on Total Aerobic bacteria (TAB) and Bile Tolerant Gram Negative (BTGN) but also speciation (Aeromonas sp., Pseudomonas sp., etc) (see summary table Table 3). This is key in the management of preventative environmental monitoring (PEM). This was also true for fungal contaminants which can also be a detriment to any grow facility.
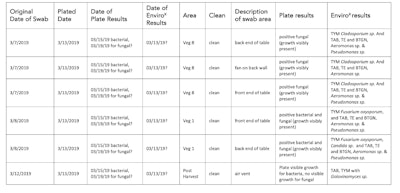 Table 3. Results summary table for swabbed presumed clean areas.
Table 3. Results summary table for swabbed presumed clean areas.
Conclusion
The sample collection at the facility took less than 30 minutes to complete. If it was done first thing in the morning, the samples could be transported to either an onsite testing facility or to a close by testing laboratory which can process the samples and obtain reportable results within 6.5 hours of receiving the samples. This translates to being able to do a complete monitoring of a site in one work shift. For large facilities which may require dozens of samples to be collected and analyzed, the overall time to result would likely take longer but still be completed within one day if they implement a split shift employment system.
Environmental monitoring of microbial species within cannabis cultivation and processing facilities can be a proactive approach towards detecting microbial contamination. Having such a capability when performed using a SMART test can provide management of a facility early warning of a contamination breech and permit implementation of a mitigation action to eradicate the contamination before it becomes systemic and prior to the possible compromising of large amounts of highly valued products.
This can be accomplished using molecular methods, including a microarray-based assay which can query for the presence of a large number of microbial species, including unculturable microbes, in parallel which can make such testing more efficient, informative and cost effective. The multiplexing capability provides a large advantage over culture methods for environmental screening. For one to use culture, enrichment is a common technique to increase the number of cells from the original sample and thus permit the splitting of the sample into a number of aliquots to accommodate the number of different culture plates necessary to identify the microbes of interest.
However, the enrichment process does introduce bias to the monitoring process due to either lack of growth of certain organisms in the enrichment medium or the interactions between various organisms which can either favor or hinder the growth of particular species (Dunbar et al., 1997). This method is also rapid compared to culture and does not require as much space since one does not have to incubate and store so many petri plates with various types of media and incubators to support the growth of so many different kinds of microbial species.
PathogenDx multiplexing capabilities offer a simple solution to the potential spreading of environmental microbial pathogens with ease of use that doesn’t require high complexity molecular testing personnel thus making it the most cost effective molecular based pathogen detection system with optimal specificity and sensitivity.
References
- M.R. Blood. “Big safety testing failure rate for California pot products.” Associated Press. https://www.apnews.com/2cb04323f9074c1ca28001693f6e2a8a (2018).
- S. Sutton. The environmental monitoring program in a GMP environment. J. GXP Compl., 14:22-30 (2010).
- Center for Food Safety and Applied Nutrition. Hazard Analysis Critical Control Point (HACCP). Retrieved April 13, 2019, from https://www.fda.gov/food/guidanceregulation/haccp/ (2018, January 29).
- D. Jansen. “Microbiological environmental Monitoring: three hurdles you need to overcome.” Contract Pharma, https://www.contractpharma.com/issues/2014-03-01/view_features/microbiologicalenvironmental-monitoring (2014).
- Kralik, P., & Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5288344/ (2017, February 02).
- McLoughlin, K. S. Microarrays for pathogen detection and analysis. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245552/ (2011, November).
- Frey,K., Bishop-Lilly, K. Next-Generation Sequencing for Pathogen Detection and Identification. Retrieved from https://www.sciencedirect.com/science/article/pii/S0580951715000136 (2015, July 31).
- J. Dunbar, S. White, and L Forney. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol., 63:1326-1331 (1997).























